Solving challenging problems using machine learning, which is the study of use of algorithms to perform regression or classification tasks without explicit instructions, is growing in popularity across a range of disciplines – including biology. In biology, many processes are not linear and one machine learning technique that can handle nonlinear relationships in data is the neural network.
A recent breakthrough in computer vision that used convolutional neural network (CNN) models led CNNs to become the standard for classification and detection tasks in imagery. Compared to other neural network models, CNNs use less pre-processing than other algorithms and the neurons only process data for their receptive field. However, most of the CNN models developed to date were trained on labelled datasets, where the classification and detection answers are known, tested in the field with multiple sensors spanning the electromagnetic spectrum, and are not small enough to be deployed on a mobile device. To deploy such models more broadly and equitably, scientists across fields are working to deploy models on mobile devices. Having a CNN model loaded on a mobile device is beneficial for rural locations that may not have reliable Internet access. Additionally, CNN models have not yet been deployed in realistic field settings when only one sensor, the phone’s camera, is available.
CIDD researchers Amanda Ramcharan, Peter McCloskey, Kelsee Baranowski, and David Hughes used a trained CNN model deployed on mobile devices to detect disease in cassava plants in Tanzania in real-time. After assembling, cleaning, and annotating a cassava disease image dataset in previous work (Ramcharan et al 2017), the CNN was trained on this dataset and deployed to the field via a TensorFlow app to detect three problems: cassava mosaic disease (CMD), cassava brown streak disease (CBSD), and green mite damage (GMD). Both mild and pronounced disease severity was assessed. Additionally, the other new challenges the CNN faced were differences in lighting and orientation of the image.
Running the CNN on the test data set resulted in a mean average precision of 94% (+/- 5.7%) and mean average recall of 67.6% (+/- 4.7%). Mean average precision dropped slightly in real world image and video, more so for mild disease (roughly 89-91% +/- 10% and 75-81.2% +/- 19-27% respectively). However, mean average recall drops dramatically in real world image and video. It drops in half for pronounced disease (approximately 39% +/- 10-20%) and down to a quarter for mild disease (approximately 16% +/- 10-11%). In general, the model performed better for CMD than CBSD and CGM. However, in some extreme cases of disease, leaf shape was altered and this impacted the algorithm. Ramcharan et al suggest machine learning algorithms trained on one stage of disease will be less accurate if plant symptoms change significantly during the course of disease.
This research provides valuable insights into the current limitations of deploying real-time machine learning algorithms into the field for plant disease detection on mobile devices. Milder symptoms in various plant diseases look similar and will need a curated, labelled dataset to be developed and used to train future models. Additionally, differences in lighting and orientation must be addressed. Despite these current challenges, the excitement of deploying accessible algorithms around the globe to help tackle major global concerns such as plant disease is merited and continues to grow. This developing technology is likely to have a large impact on agriculture and will improve lives globally as it becomes a staple of future plant care.
Synopsis written by Catherine Herzog
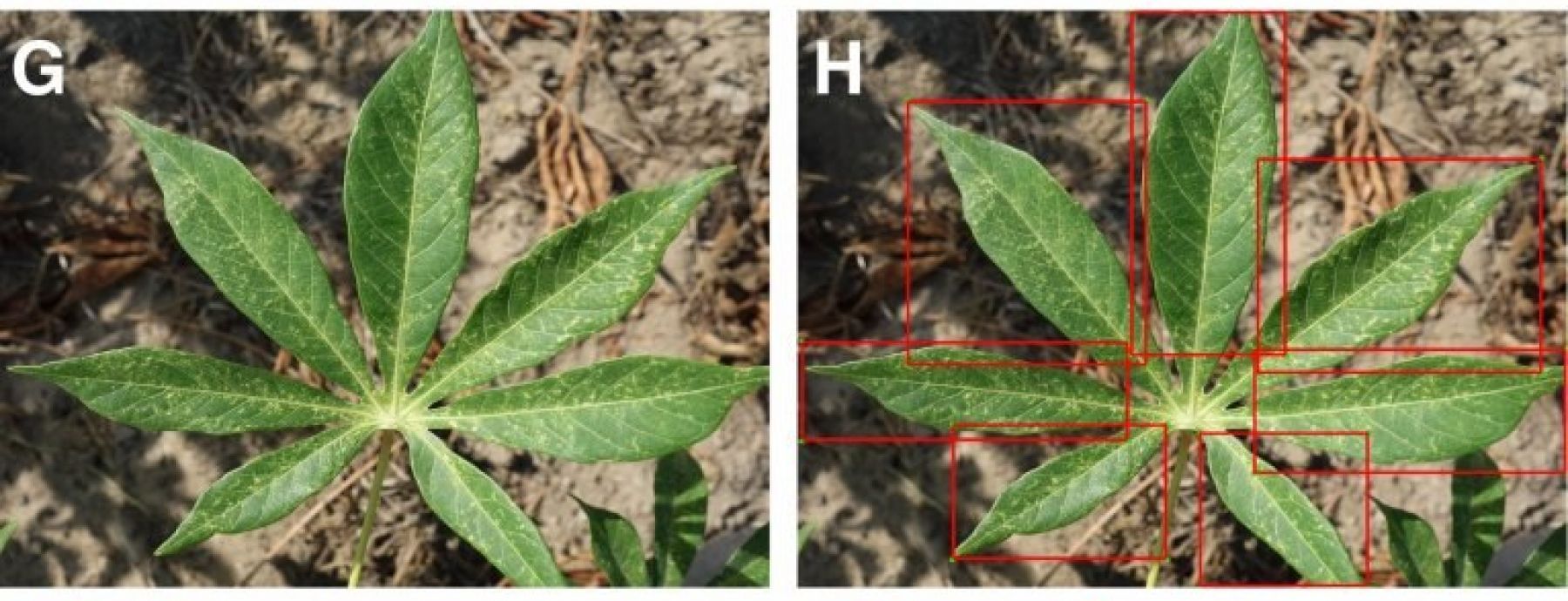
Image caption: Examples of training images from 7 classes with leaflet annotations. Classes are (A,B) Healthy, (C,D) Brown streak disease, (E,F) Mosaic disease, (G,H) Green mite damage, (I,J) Red mite damage, (K,L) Brown leaf spot and (M,N) Nutrient Deficiency.
More information on PlantVillage can be found here and by watching the video below.
Written By: Amanda Ramcharan, Peter McCloskey, Kelsee Baranowski, Neema Mbilinyi, Latifa Mrisho, Mathias Ndalahwa, James Legg, David Hughes
Paper Url: https://www.frontiersin.org/articles/10.3389/fpls.2019.00272/full
Journal: Frontiers in Plant Science